| Distinguishing between normal, cancerous, and metastatic cells is a major hurdle for the early detection of cancer. Bajaj, et. al. describe a detection approach based on selective noncovalent interactions between cell surface elements and functionalized nanoparticle sensors that does not require a priori knowledge of specific biomarkers. |
Reviewed by Jeff Morse, PhD., National Nanomanufacturing Network
Distinguishing between normal, cancerous, and metastatic cells is a major hurdle for the early detection of cancer, as each cell type has a unique molecular signature which includes both intracellular and cell surface biomarkers. The earlier these signatures can be detected, the more effectively they can be treated. A range of techniques have been studied to this end. While some approaches require a priori knowledge of specific mutations in DNA/RNA for both intracellular and cell surface biomarkers, others, such as array-based antibodies, have insufficient sensitivity and specificity to differentiate between normal, cancerous, and metastatic cells.
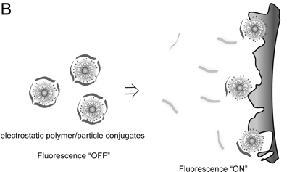
The detection system is based on conjugates of three related cationic gold nanoparticle structures and the poly(para-phenyleneethynylene) (PPE) polymer PPE-CO2. For these particular conjugates, the nanoparticle quenches the fluorescence of the polymer. When the nanoparticle-polymer complexes are incubated with cells, competitive binding between the cell surfaces and the nanoparticle-polymer complexes occur. As the nanoparticles interact with the cell surface, the polymer fluorophore is displaced from the nanoparticle-polymer complex, thereby generating a fluorescent response. As a result, the response can either decrease fluorescence through displacement of the polymer fluorophores, or increase fluorescence though quenching of residual PPE-CO2 flourescence by the cell surface. Consequently, the nanoparticles can be designed to have specific affinities to a different cell surfaces depending on cell membrane composition.
The authors demonstrated the utility of this approach by differentiating between several normal and cancerous mammalian cell lines, thereby providing the basis for a rapid and effective array-based methodology for the detection of diseased tissue. In the results, full differentiation between normal and cancerous cell lines was achieved using only three nanoparticle-polymer complexes demonstrating the benefit of this approach as a potential diagnostic capability. Furthermore, these nanoparticle-polymer systems have the potential to provide new understanding of physiochemical changes occurring on cell surfaces in various disease states, enabling new areas of fundamental research into advanced biodiagnostics and surface science involving cells.
Image source: Bajaj A, et.al. 2009. Detection and Differentiation of Normal, Cancerous, and Metastatic Cells Using Nanoparticle-Polymer Sensor Arrays.PNAS 106(27): 10912-10916. DOI: 10.1073/pnas.0900975106. Copyright PNAS 2009.
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported.
