| Kershner, et. al., report a synergistic top down and bottom up process for creating self-assembled nanostructures of DNA origami. |
Reviewed by Jeff Morse, PhD, National Nanomanufacturing Network
DNA nanostructures have been investigated as a means to organize functional nanomaterials with high resolution. For example, DNA origami—long, folded single DNA strands fixed in a variety of shapes by shorter DNA strands—can generate 6nm resolution patterns at specific binding sites. Conceivably, these binding sites could enable a complex arrangement of nanostructures to form an electronic or optical device or circuit at scales unobtainable by other nanofabrication methods. While this may lead to some intriguing possibilities for future generations of nanoelectronics, the challenge thus far has been to control the positioning and orientation of the DNA origami adsorbed to a given substrate from solution.
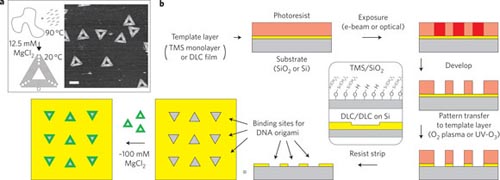
Recently in Nature Nanotechnology, Kershner et. al. report an approach to forming controlled arrangements and orientations of triangular DNA origami. Using both diamond like carbon (DLC) and silicon wafers coated with a monolayer of trimethylsilyl (TMS), the authors first coat the surface with photoresist, then pattern the resist using electron beam lithography in the same shape and size as their DNA origami—in this case triangles with 127nm sides. The pattern is then transferred onto the template layer using an oxidative plasma etch step. When the photoresist is removed, the remaining patterned templates are hydrophilic and exhibit high selectivity for binding DNA origami. The templated substrate is then immersed in a solution of nanomolar concentration origami with approximately 100 mM MgCl2 in a buffer of Tris-acetate-EDTA (TAE). The MgCl2 provides the high concentration of divalent cations to facilitate the adsorption and binding of the DNA origami from solution. The templated substrates are immersed in the buffer solution containing DNA origami for periods ranging from several minutes to over an hour.
Subsequent imaging and assessment of the origami adsorbed to the substrates reveals excellent selectivity compared to an untreated surface, with >70% of the sites having alignment dispersions of +/-10-20°. Further variation of the size and shape of the templates demonstrates that multiple origami could assemble within a template thereby providing further versatility to the process.
While significant research remains to reduce the orientation dispersion and improve the deposition rate of the origami, this work is a good illustration of synergistic top down and bottom up processes for creating self-assembled nanostructures. Future investigations for this work may consider different size and shaped origami, as well as different surfaces on which these nanostructures can be adsorbed. Ultimately this method could provide a means to integrate specific nanostructures within the DNA origami scaffold and integrate these elements with larger circuits or devices.
Image reprinted by permission from Macmillan Publishers Ltd:Kershner RJ, et. al. 2009. Placement and Orientation of Individual DNA Shapes on Lithographically Patterned Surfaces. Nature Nanotechnology 4:557-561, copyright 2009.
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported.
